New BioRχiv pre-print
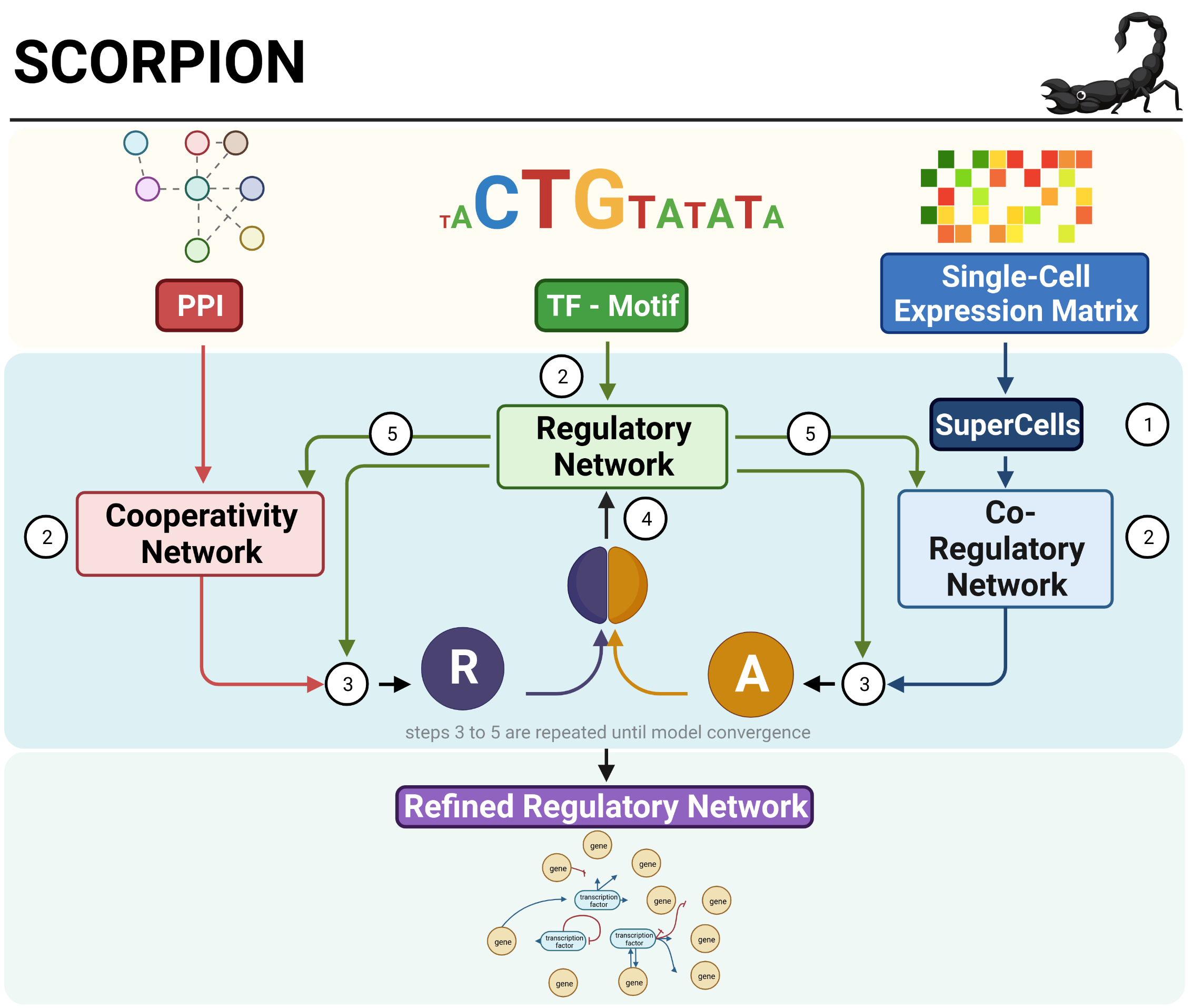
We have posted a new BioRχiv pre-print by Daniel. Daniel developed SCORPION, or Single Cell Oriented Reconstruction of PANDA-based Individually Optimized Networks, an approach to reconstruct gene regulatory networks based on single cell transcriptomic data.
SCORPION combines the concept of SuperCells with PANDA’s message passing framework. This reduces sample size and de-sparsifies the single cell expression data to render network modeling possible on large single cell datasets. Daniel benchmarked SCORPION against 12 other network modeling algorithm and found it to model accurate networks, while also being the second fastest approach. We further tested SCORPION in single cell data from epithelial cells from mouse intestines, and found it captured differential targeting by overexpressed or knockouts of transcription factors. In colon cancer, SCORPION captured known dysregulated pathways. Finally, we identified differential regulation between left- and right-sided colon cancer that may explain differences in patient survival and could potentially have implications for personalized treatment of the disease.
More information on the pre-print can be found here.