New publication
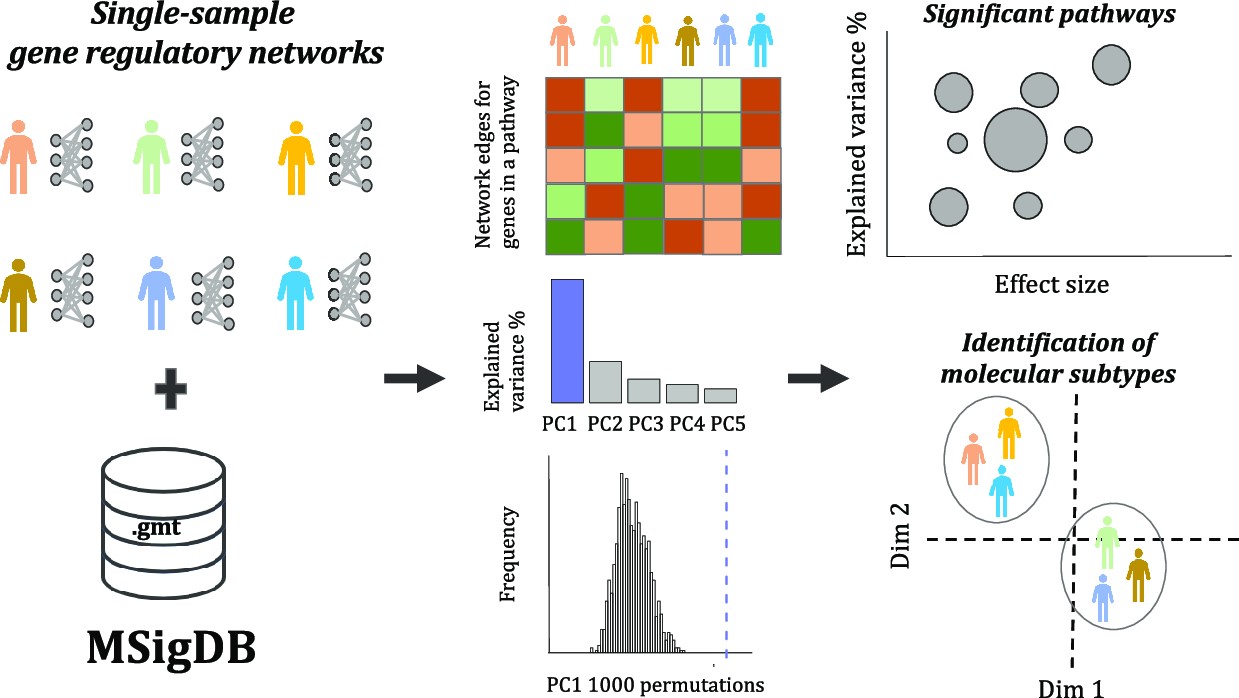
We recently published our latest work, led by Tatiana, in NAR Cancer. Tatiana developed a computational approach, called PORCUPINE, to map heterogeneity in individual patient gene regulatory networks and applied it to study leiomyosarcoma, an aggressive cancer that can develop in smooth muscle tissue.
- PORCUPINE combines PCA with permutations to identify pathways contributing to regulatory heterogeneity in a patient population. Importantly, instead of identifying discrete subtypes, it can map subtle gradients of heterogeneity not detected by standard subtyping methods. Applying the method to two leiomyosarcoma data sets, we identified 37 heterogeneously regulated pathways, incl. targetable pathways such as E2F signaling. These findings were independent of mutations and could thus be a new way of stratifying patients for personalized medicine. Finally, we found that heterogeneous regulation is generally associated with open chromatin—indicating that genes located in open chromatin are more likely to be regulated by different sets of TFs—and that differential accessibility may explain subtle differences in regulation.
- In summary, we uncovered patterns of inter-patient heterogeneity at the level of transcriptional regulation, and identified genes and pathways that may represent therapeutic entry points in leiomyosarcoma.
- For more information, please see the publications section.