New BioRχiv pre-print
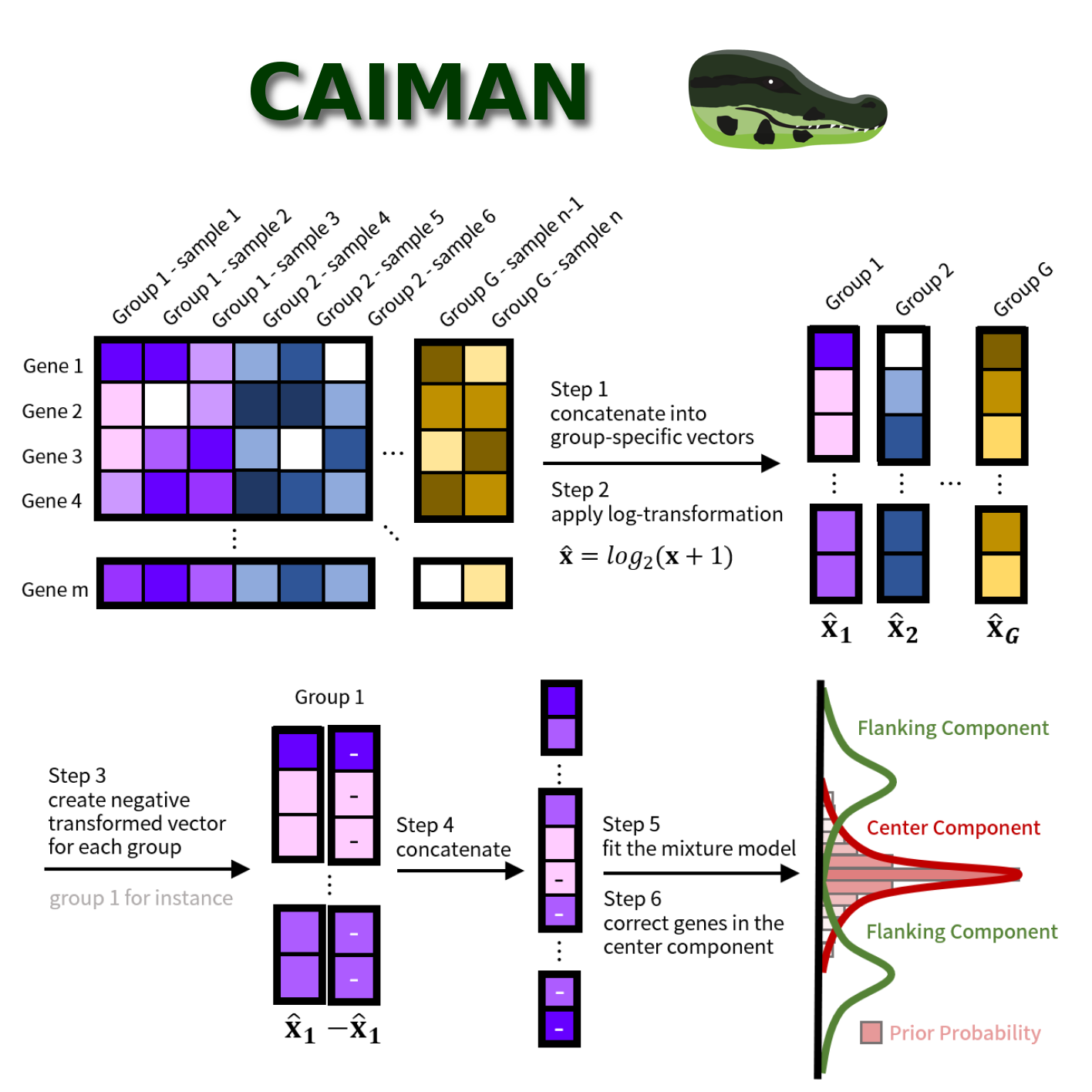
We have a new BioRχiv pre-print by Ping-Han! Ping-Han found that certain normalization methods, in specific quantile-based methods, can introduce false positive associations in co-expression measurements. However, there methods can be very powerful. Smooth quantile normalization, for example, allows one to normalize heterogeneous data, while keeping global expression differences between different subgroups of samples. Ping-Han therefore developed a new algorithm, called CAIMAN, to correct for these false-positive associations. Importantly, with CAIMAN, there is not need to filter out lowly expressed genes (that are prone to form false positive associations) based on arbitrary thresholds. Thus, CAIMAN allows for more precise comparative network analysis in large-scale heterogeneous data. More information on the pre-print can be found here.